Back to agmial Main Page
User Guide
Artemis is a genome viewer developped by the
Sanger Institute that allows visualization of sequence features and
the results of analyses within the context of the sequence, and its
six-frame translation. Artemis has been modified to access Agmial
servers and to display features like MuGeN (Box coloration, Features
shapes).
This page only describes the modifications made to the standard
version of artemis. For a complete user guide of artemis, see Sanger site.
System Requirements
Artemis will run on any machine that has a recent version of Java.
Agmial access
Artemis Launch
(Nothing happens ? See Faq). If needed, select javaws (JNLPFile under Windows) as the default program to open jnlp files. ( See FAQ)
Artemis Loading
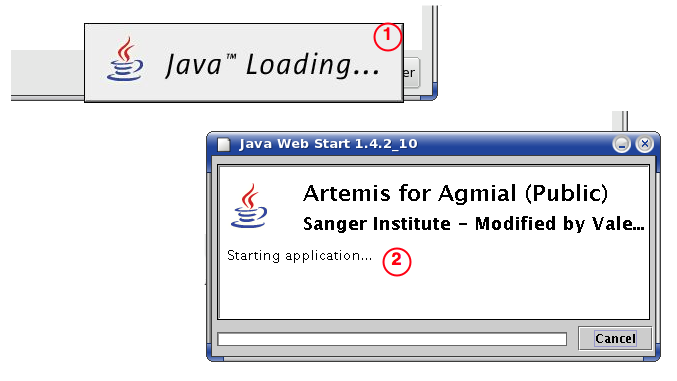
Then, the application is loaded (or updated) from our servers [2].
Security Constraints
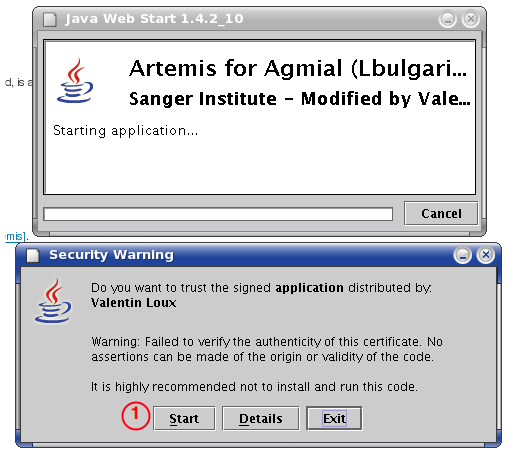
Inra Agmial Access
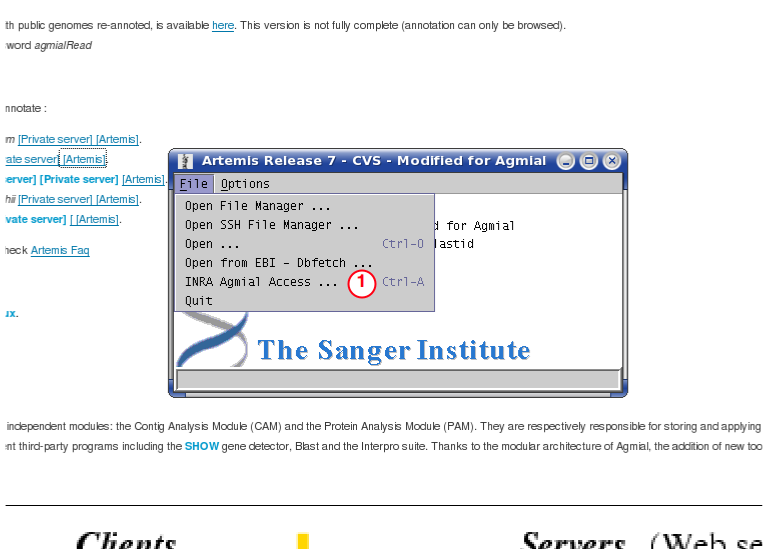
Authentification
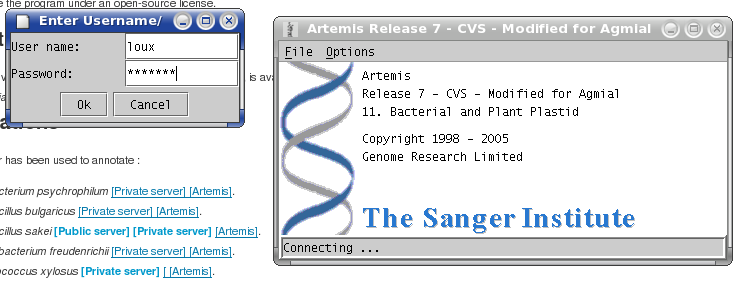
Agmial Project Dialog
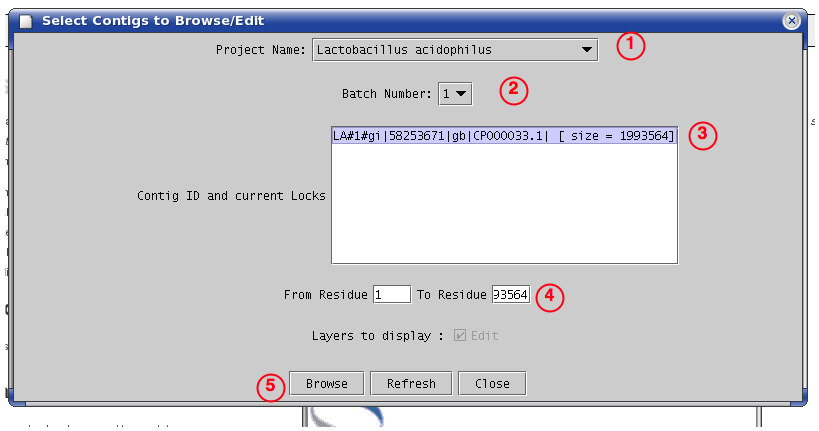
Then click on "Browse"[5].
Artemis
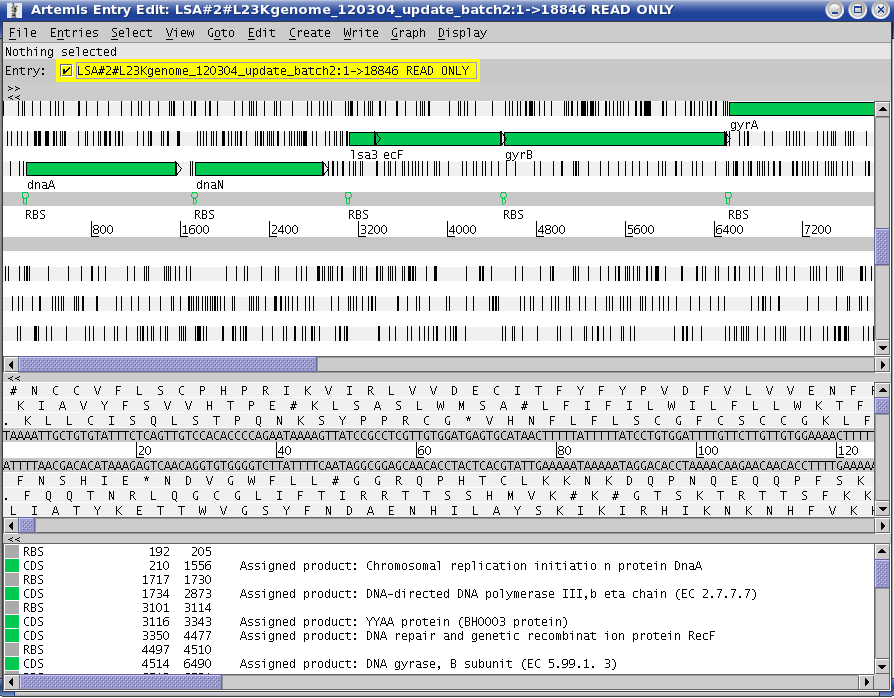
You are now able to navigate through the genome. A click on a CDS feature open its PAM page (resuming results and annotations) in the default web browser (Not the right one ? See FAQ)
top
Faq
Question: When I click on the artemis link, nothing happens
First of all, verify at this address that you
have a valid Java software installation. You should - at least - have
Java 1.4.2 . If not, follow the instructions on thoses pages to
install it.
Question: When I click on the artemis link, my browser ask me what I want to do with the file.
You have to associate the type of files Java WebStart use to define deployable applications (JNLP file) with the appropriate programm (javaws for Linux, JNLPFiles for Windows).
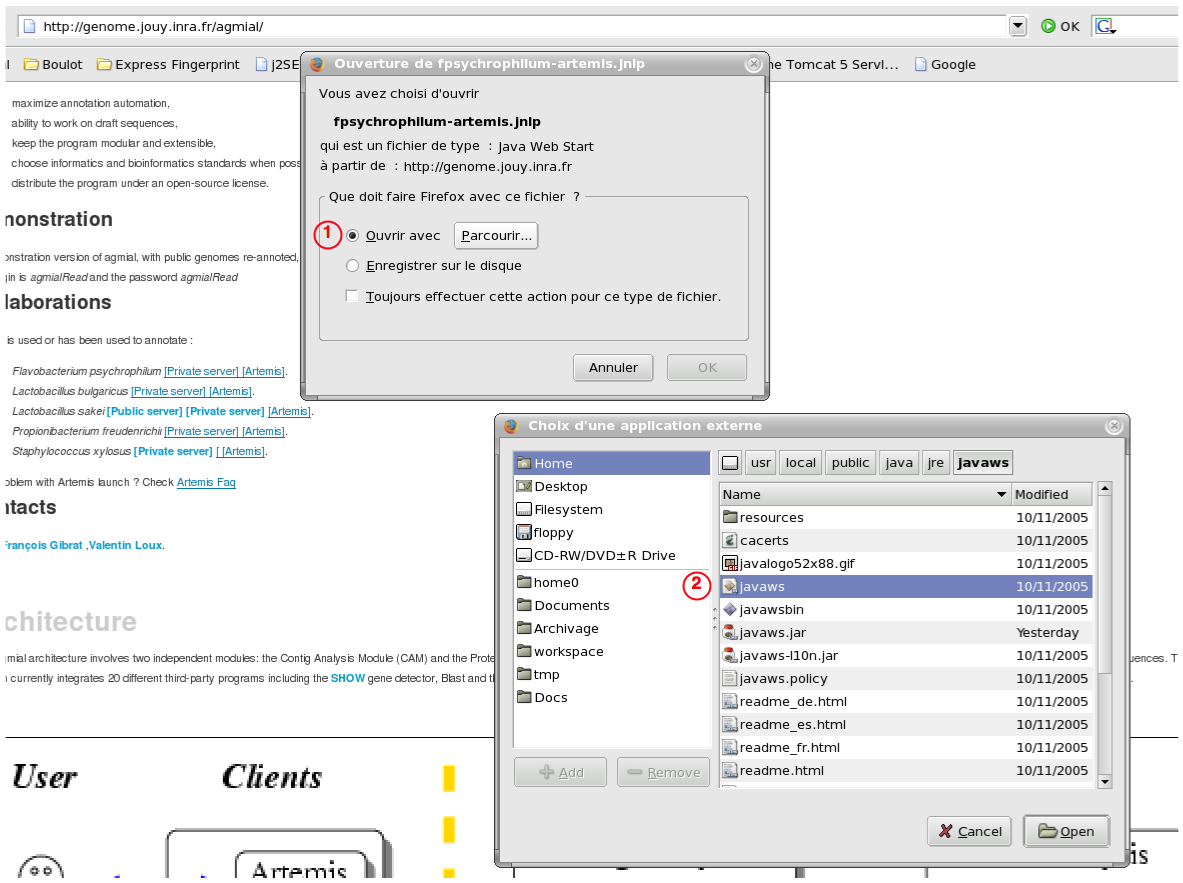
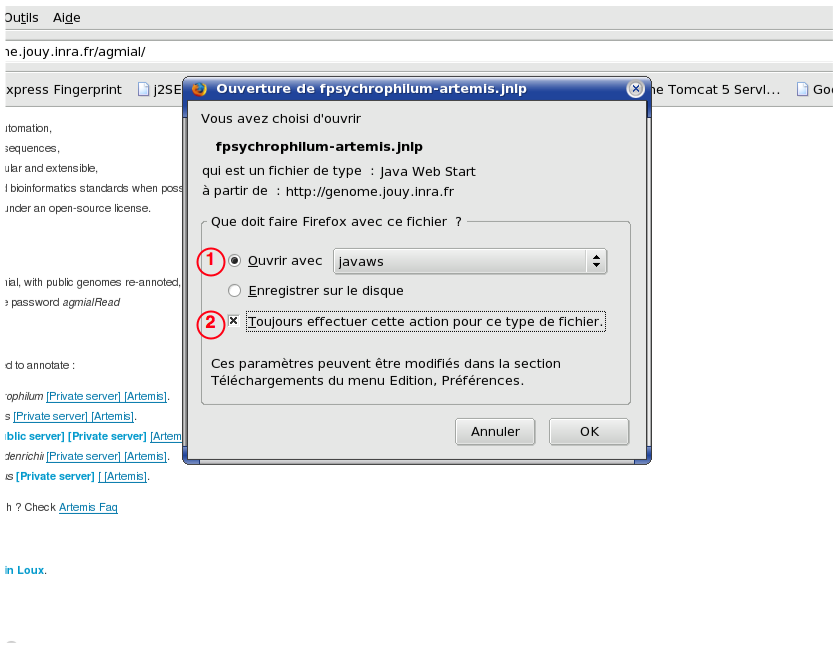
. By clicking on the "Always perform this action for that kind of file" checkbox [2], you save you preferences.
Question: When I click on a feature (with Agmial Protein Browsing enabled), artemis launches Internet Explorer© instead of my prefered browser (Firefox, for example)
Artemis is set to lauch your default browser. How to change your default browser
