| Supplementary data of the associated publication Start Page |
|
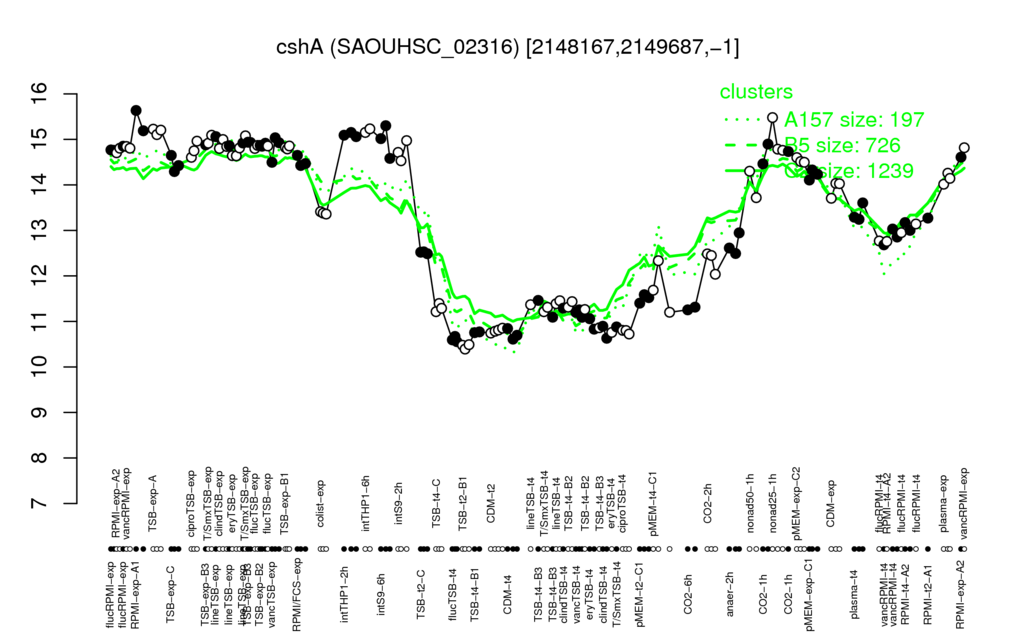
| RPMI-exp-A1 | intTHP1-6h | TSB-exp-A | intTHP1-2h | T/SmxTSB-exp |
| 15.4 | 15.2 | 15.2 | 15.1 | 15 |
| TSB-t2-B1 | flucTSB-t4 | CDM-t4 | TSB-t4-B1 | ciproTSB-t4 |
| 10.5 | 10.6 | 10.7 | 10.8 | 10.8 |
| S894 | rho | S183 | rplL | S913 | SAOUHSC_01392 | rpsD | pyrG | SAOUHSC_01077 | SAOUHSC_01756 |
| 0.98 | 0.97 | 0.96 | 0.96 | 0.96 | 0.96 | 0.95 | 0.95 | 0.95 | 0.95 |
| pnbA | SAOUHSC_02244 | SAOUHSC_02928 | S1065 | hutG | SAOUHSC_00318 | SAOUHSC_00320 | SAOUHSC_00319 | S957 | SAOUHSC_02581 |
| -0.95 | -0.94 | -0.93 | -0.93 | -0.93 | -0.93 | -0.93 | -0.93 | -0.92 | -0.92 |
