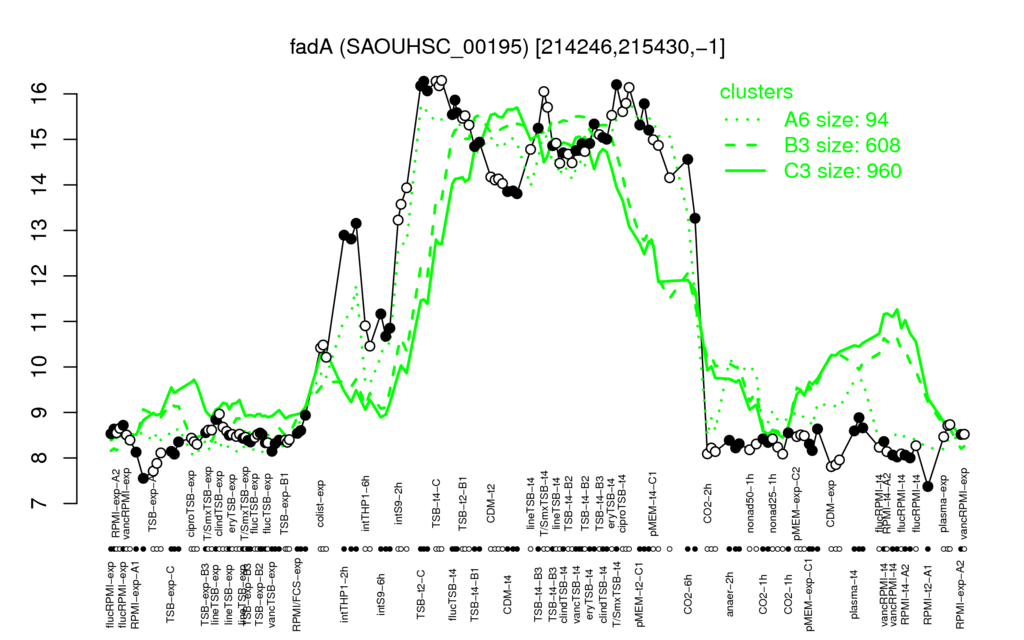
| Supplementary data of the associated publication Start Page |
|
| TSB-t4-C | TSB-t2-C | T/SmxTSB-t4 | ciproTSB-t4 | flucTSB-t4 |
| 16.2 | 16.2 | 16 | 15.8 | 15.7 |
| RPMI-t2-A1 | RPMI-exp-A1 | CDM-exp | TSB-exp-A | RPMI-t4-A2 |
| 7.37 | 7.84 | 7.87 | 7.9 | 8.07 |
| fadB | S64 | SAOUHSC_00194 | fadD | S63 | S65 | acsA | fadE | rocA | murQ |
| 1 | 1 | 0.99 | 0.99 | 0.99 | 0.99 | 0.99 | 0.99 | 0.99 | 0.98 |
| S279 | SAOUHSC_00190 | gapR | trpE | S322 | SAOUHSC_00462 | guaA | SAOUHSC_00607 | SAOUHSC_02372 | S120 |
| -0.91 | -0.9 | -0.9 | -0.9 | -0.9 | -0.89 | -0.89 | -0.89 | -0.88 | -0.88 |
