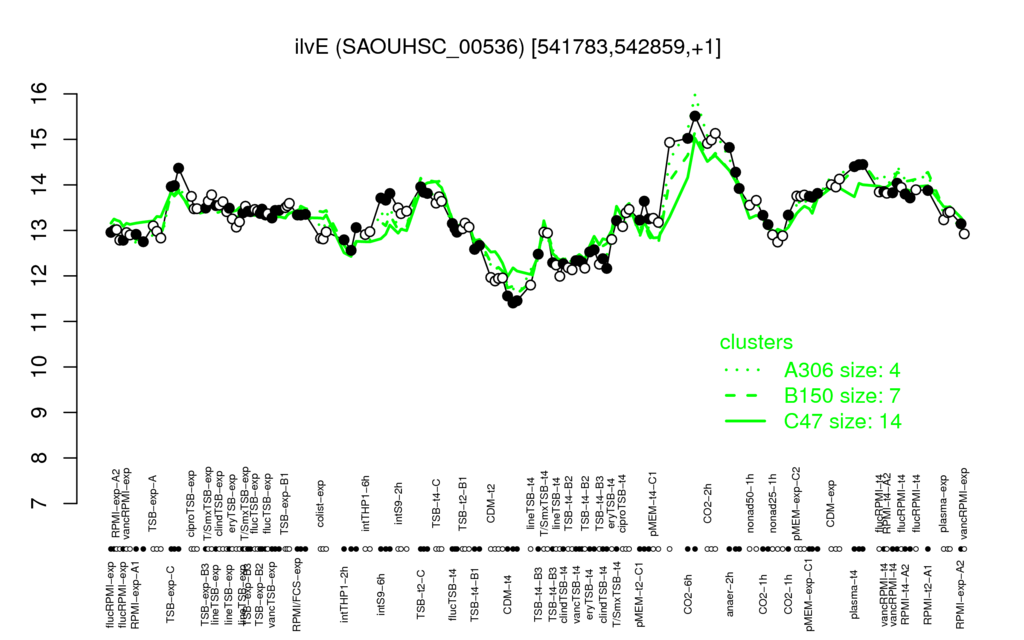
| Supplementary data of the associated publication Start Page |
|
| CO2-6h | CO2-2h | plasma-t4 | anaer-2h | TSB-exp-C |
| 15.3 | 15 | 14.4 | 14.3 | 14.1 |
| CDM-t4 | CDM-t2 | lineTSB-t4 | TSB-t4-B2 | clindTSB-t4 |
| 11.5 | 11.9 | 12 | 12.2 | 12.3 |
| SAOUHSC_00537 | S194 | SAOUHSC_00538 | SAOUHSC_01868 | SAOUHSC_00989 | relA | nrdE | SAOUHSC_02576 | SAOUHSC_01895 | nrdF |
| 0.95 | 0.95 | 0.85 | 0.84 | 0.81 | 0.8 | 0.8 | 0.78 | 0.78 | 0.78 |
| S313 | dinP | SAOUHSC_01468 | SAOUHSC_02322 | SAOUHSC_02321 | S830 | SAOUHSC_02275 | S312 | dus | SAOUHSC_01507 |
| -0.77 | -0.77 | -0.77 | -0.76 | -0.76 | -0.76 | -0.74 | -0.74 | -0.73 | -0.73 |
