| Supplementary data of the associated publication Start Page |
|
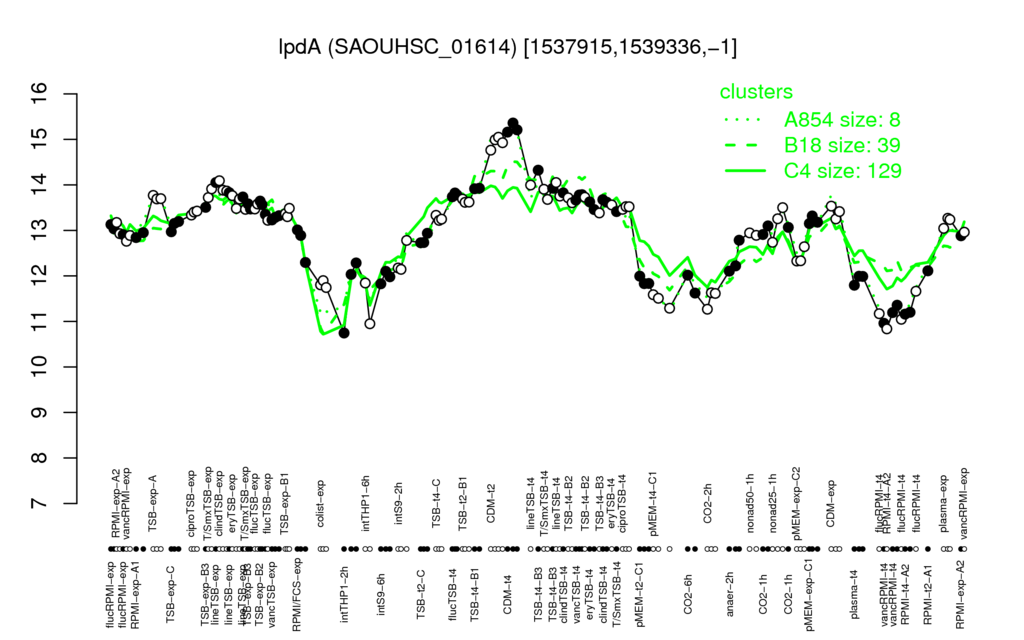
| CDM-t4 | CDM-t2 | clindTSB-exp | lineTSB-t4 | TSB-t4-B1 |
| 15.2 | 14.9 | 13.9 | 13.9 | 13.9 |
| RPMI-t4-A2 | vancRPMI-t4 | flucRPMI-t4 | intTHP1-6h | pMEM-t4-C1 |
| 11.1 | 11.2 | 11.3 | 11.4 | 11.5 |
| bfmBAB | bfmBAA | S646 | bmfBB | recN | S645 | gyrB | gyrA | argR | dnaJ |
| 0.98 | 0.98 | 0.97 | 0.94 | 0.91 | 0.9 | 0.89 | 0.89 | 0.87 | 0.84 |
| SAOUHSC_00828 | SAOUHSC_02129 | SAOUHSC_01062 | SAOUHSC_02923 | S845 | SAOUHSC_02888 | SAOUHSC_00996 | S264 | S1149 | ilvD |
| -0.88 | -0.82 | -0.81 | -0.79 | -0.78 | -0.78 | -0.77 | -0.77 | -0.77 | -0.77 |
