| Supplementary data of the associated publication Start Page |
|
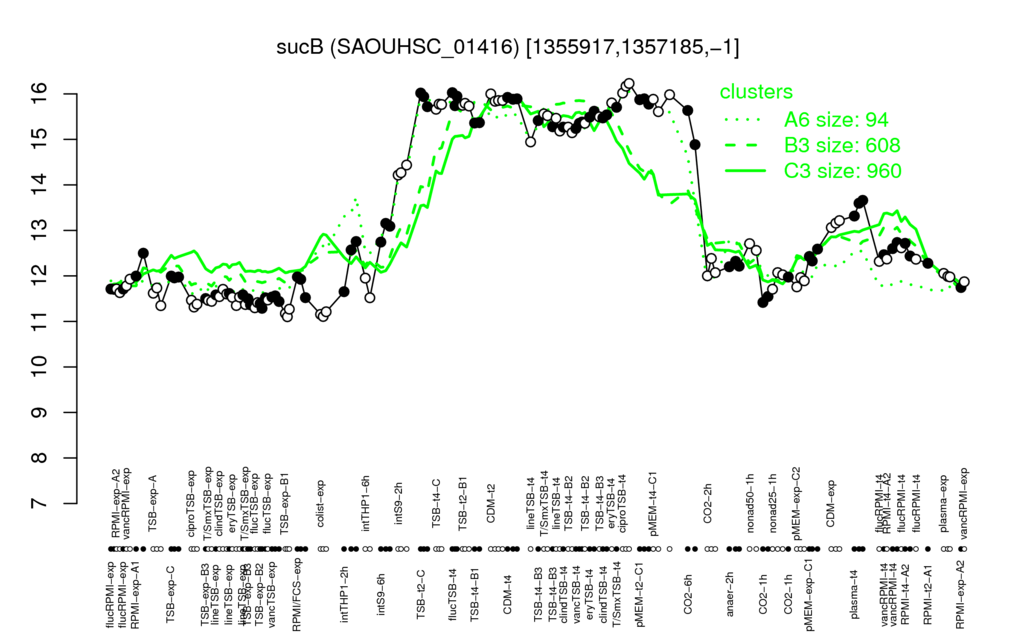
| ciproTSB-t4 | flucTSB-t4 | CDM-t4 | TSB-t2-C | CDM-t2 |
| 16.1 | 15.9 | 15.9 | 15.9 | 15.9 |
| colist-exp | TSB-exp-B1 | ciproTSB-exp | flucTSB-exp | TSB-exp-B2 |
| 11.2 | 11.2 | 11.4 | 11.4 | 11.4 |
| sucA | S593 | rocF | gudB | fadE | pckA | SAOUHSC_00156 | murQ | S65 | fadD |
| 0.99 | 0.98 | 0.96 | 0.96 | 0.96 | 0.95 | 0.95 | 0.95 | 0.95 | 0.95 |
| S970 | glcU | braB | S971 | S279 | S568 | SAOUHSC_01338 | alsT | proS | SAOUHSC_A02450 |
| -0.95 | -0.94 | -0.93 | -0.93 | -0.93 | -0.93 | -0.92 | -0.91 | -0.91 | -0.91 |
