| Supplementary data of the associated publication Start Page |
|
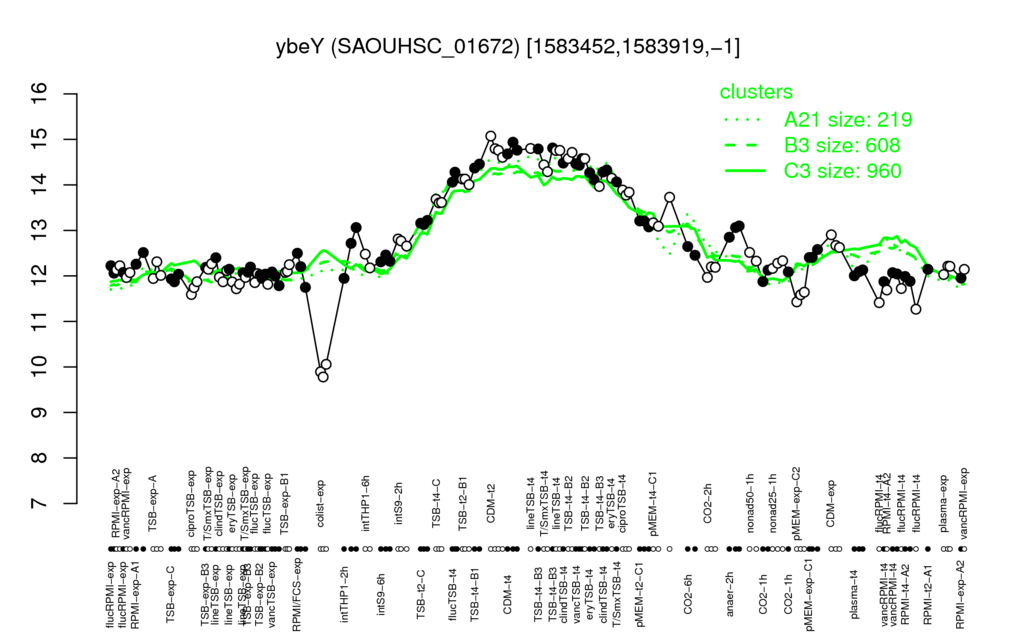
| CDM-t2 | CDM-t4 | lineTSB-t4 | TSB-t4-B2 | TSB-t4-B3 |
| 14.8 | 14.8 | 14.8 | 14.6 | 14.5 |
| colist-exp | flucRPMI-t4 | pMEM-exp-C2 | ciproTSB-exp | eryTSB-exp |
| 9.91 | 11.5 | 11.6 | 11.7 | 11.8 |
| dgkA | phoH | S660 | era | cdd | SAOUHSC_01650 | tagB | SAOUHSC_00163 | SAOUHSC_00164 | epiF |
| 0.99 | 0.98 | 0.98 | 0.98 | 0.98 | 0.95 | 0.95 | 0.94 | 0.93 | 0.92 |
| S1101 | SAOUHSC_02823 | SAOUHSC_00433 | SAOUHSC_02104 | SAOUHSC_00959 | SAOUHSC_02599 | ltrA | rpsP | crcB1 | SAOUHSC_00863 |
| -0.96 | -0.95 | -0.94 | -0.93 | -0.93 | -0.93 | -0.93 | -0.92 | -0.92 | -0.92 |
