| Supplementary data of the associated publication Start Page |
|
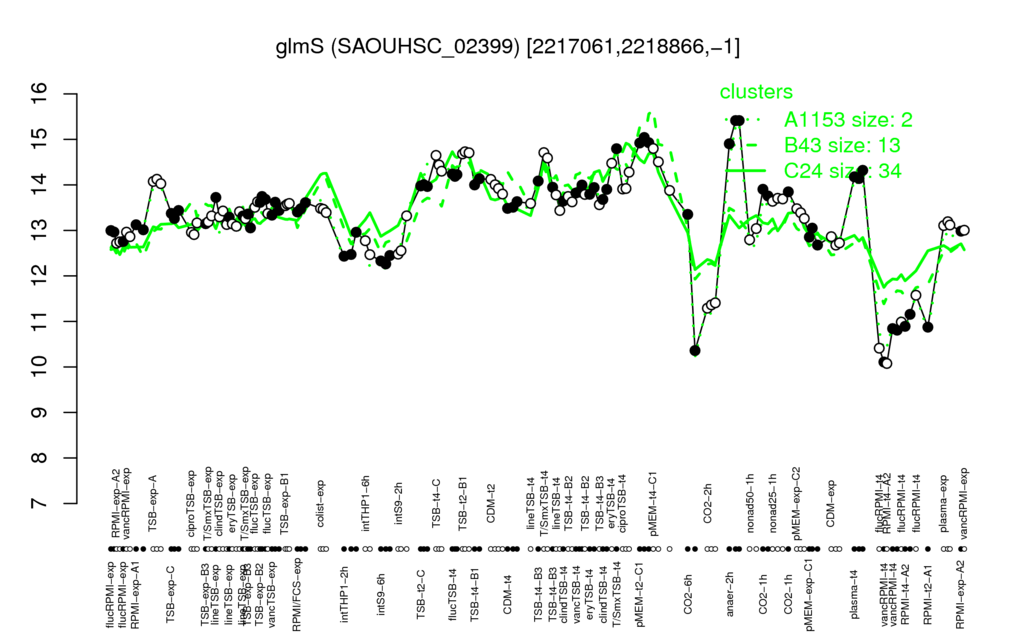
| anaer-2h | pMEM-t2-C1 | TSB-t2-B1 | T/SmxTSB-t4 | TSB-t4-C |
| 15.2 | 15 | 14.7 | 14.7 | 14.5 |
| vancRPMI-t4 | RPMI-t4-A2 | RPMI-t2-A1 | flucRPMI-t4 | CO2-2h |
| 10.6 | 10.7 | 10.9 | 11 | 11.4 |
| S936 | S619 | scpA | moaB | moeB | S980 | S981 | SAOUHSC_00096 | gyrA | gyrB |
| 0.97 | 0.75 | 0.74 | 0.73 | 0.73 | 0.73 | 0.72 | 0.72 | 0.72 | 0.71 |
| S385 | SAOUHSC_01899 | S597 | fhuB | hisZ | fhuG | hisB | SAOUHSC_03012 | S345 | SAOUHSC_02595 |
| -0.8 | -0.76 | -0.75 | -0.74 | -0.74 | -0.74 | -0.74 | -0.74 | -0.73 | -0.73 |
