| Supplementary data of the associated publication Start Page |
|
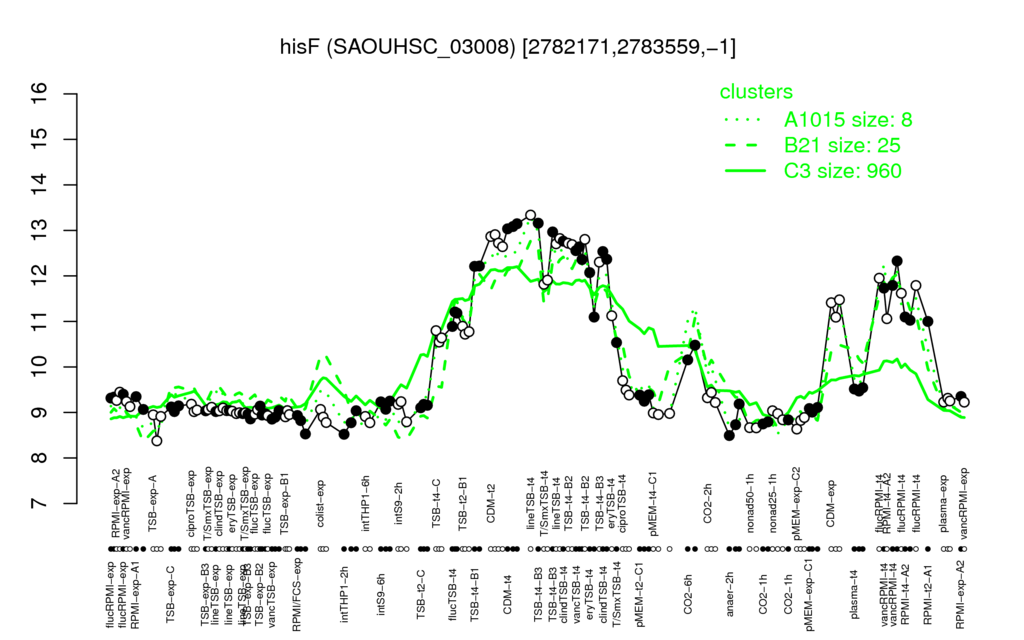
| CDM-t4 | lineTSB-t4 | TSB-t4-B3 | CDM-t2 | TSB-t4-B2 |
| 13.1 | 13 | 12.8 | 12.8 | 12.7 |
| nonad50-1h | TSB-exp-A | RPMI/FCS-exp | intTHP1-2h | pMEM-exp-C2 |
| 8.66 | 8.74 | 8.76 | 8.78 | 8.78 |
| S1178 | hisA | hisH | S804 | SAOUHSC_02138 | sspC | SAOUHSC_00820 | S339 | SAOUHSC_00854 | SAOUHSC_00810 |
| 0.99 | 0.99 | 0.94 | 0.92 | 0.92 | 0.92 | 0.9 | 0.9 | 0.9 | 0.9 |
| SAOUHSC_01837 | S189 | mqo2 | SAOUHSC_00526 | S726 | S187 | rpsL | tuf | S188 | rpsG |
| -0.92 | -0.91 | -0.91 | -0.9 | -0.9 | -0.9 | -0.89 | -0.89 | -0.89 | -0.89 |
