| Supplementary data of the associated publication Start Page |
|
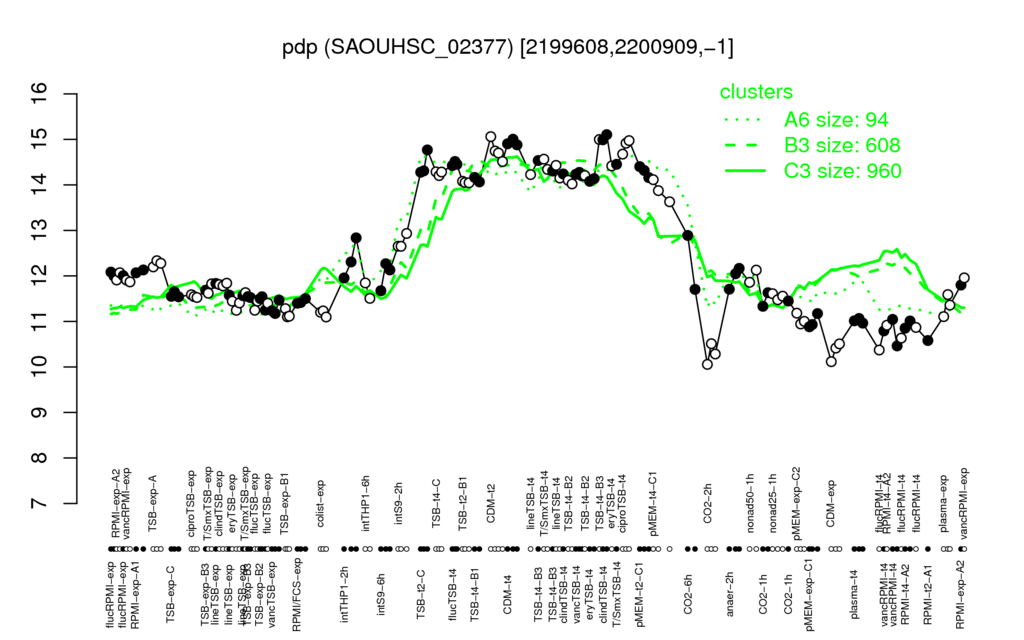
| CDM-t4 | ciproTSB-t4 | clindTSB-t4 | CDM-t2 | TSB-t4-B3 |
| 14.9 | 14.9 | 14.8 | 14.8 | 14.6 |
| CO2-2h | CDM-exp | RPMI-t2-A1 | flucRPMI-t4 | vancRPMI-t4 |
| 10.3 | 10.3 | 10.6 | 10.6 | 10.8 |
| S919 | deoC2 | SAOUHSC_02376 | S920 | SAOUHSC_00655 | SAOUHSC_00656 | SAOUHSC_00658 | murQ | SAOUHSC_00156 | S456 |
| 0.98 | 0.97 | 0.96 | 0.96 | 0.95 | 0.95 | 0.94 | 0.94 | 0.94 | 0.94 |
| S581 | SAOUHSC_02372 | guaA | SAOUHSC_00190 | S120 | S915 | xpt | SAOUHSC_02689 | SAOUHSC_00531 | guaB |
| -0.94 | -0.93 | -0.93 | -0.93 | -0.92 | -0.92 | -0.91 | -0.91 | -0.91 | -0.91 |
