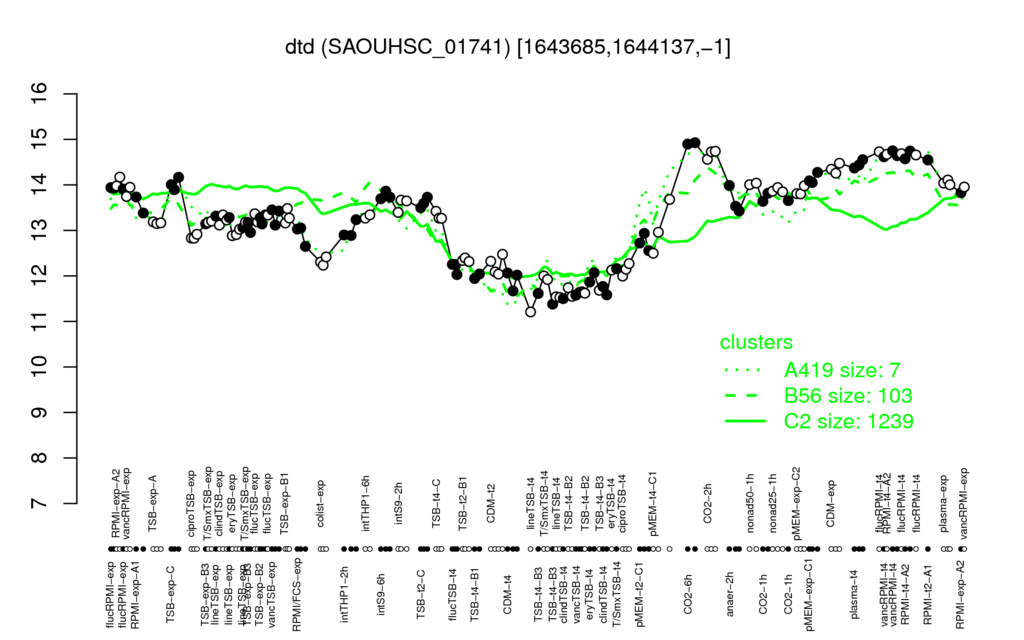
| Supplementary data of the associated publication Start Page |
|
| CO2-6h | flucRPMI-t4 | CO2-2h | vancRPMI-t4 | RPMI-t4-A2 |
| 14.9 | 14.7 | 14.7 | 14.7 | 14.7 |
| lineTSB-t4 | TSB-t4-B3 | clindTSB-t4 | vancTSB-t4 | TSB-t4-B2 |
| 11.4 | 11.6 | 11.6 | 11.6 | 11.6 |
| relA | nrdF | nrdE | SAOUHSC_00711 | purR | SAOUHSC_01908 | S293 | S275 | SAOUHSC_00995 | SAOUHSC_01868 |
| 0.93 | 0.89 | 0.89 | 0.87 | 0.87 | 0.87 | 0.86 | 0.86 | 0.86 | 0.86 |
| dinP | S830 | SAOUHSC_00099 | S312 | S265 | SAOUHSC_02275 | SAOUHSC_01507 | SAOUHSC_02727 | SAOUHSC_00133 | SAOUHSC_00041 |
| -0.91 | -0.9 | -0.89 | -0.87 | -0.86 | -0.85 | -0.85 | -0.85 | -0.84 | -0.84 |
