| Supplementary data of the associated publication Start Page |
|
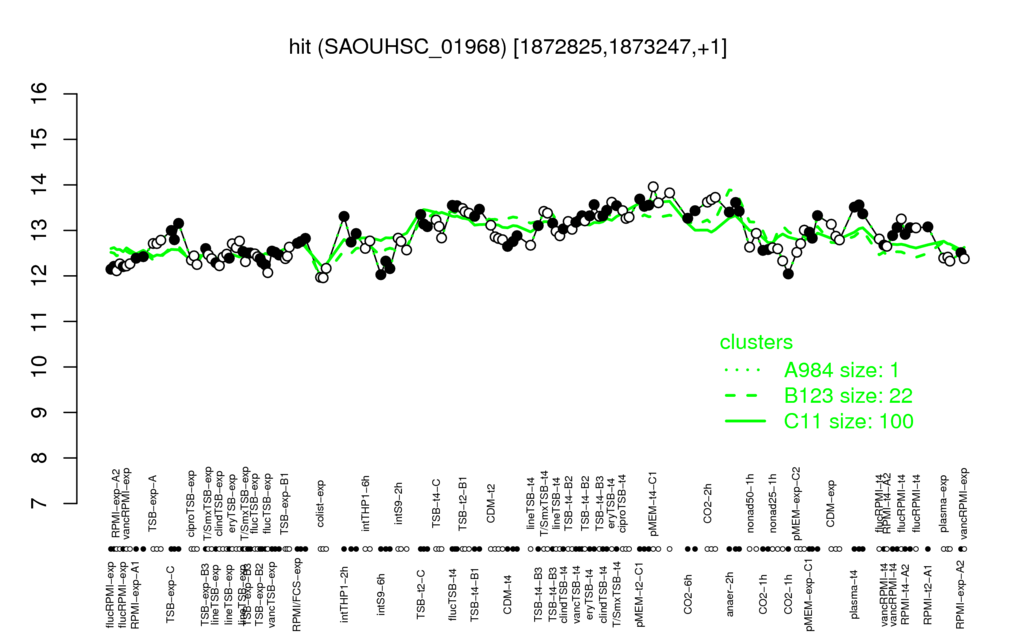
| pMEM-t4-C1 | CO2-2h | pMEM-t2-C1 | flucTSB-t4 | eryTSB-t4 |
| 13.8 | 13.7 | 13.6 | 13.5 | 13.5 |
| colist-exp | intS9-6h | flucRPMI-exp | vancRPMI-exp | RPMI-exp-A2 |
| 12 | 12.2 | 12.2 | 12.3 | 12.3 |
| rpsA | SAOUHSC_02650 | S1029 | katA | S881 | S1171 | SAOUHSC_02150 | orfX | SAOUHSC_02466 | S11 |
| 0.79 | 0.77 | 0.77 | 0.76 | 0.76 | 0.76 | 0.76 | 0.75 | 0.75 | 0.75 |
| SAOUHSC_00418 | aspS | S945 | S648 | S478 | S472 | S166 | S133 | SAOUHSC_02426 | S692 |
| -0.79 | -0.77 | -0.77 | -0.77 | -0.76 | -0.76 | -0.76 | -0.75 | -0.75 | -0.75 |
