| Supplementary data of the associated publication Start Page |
|
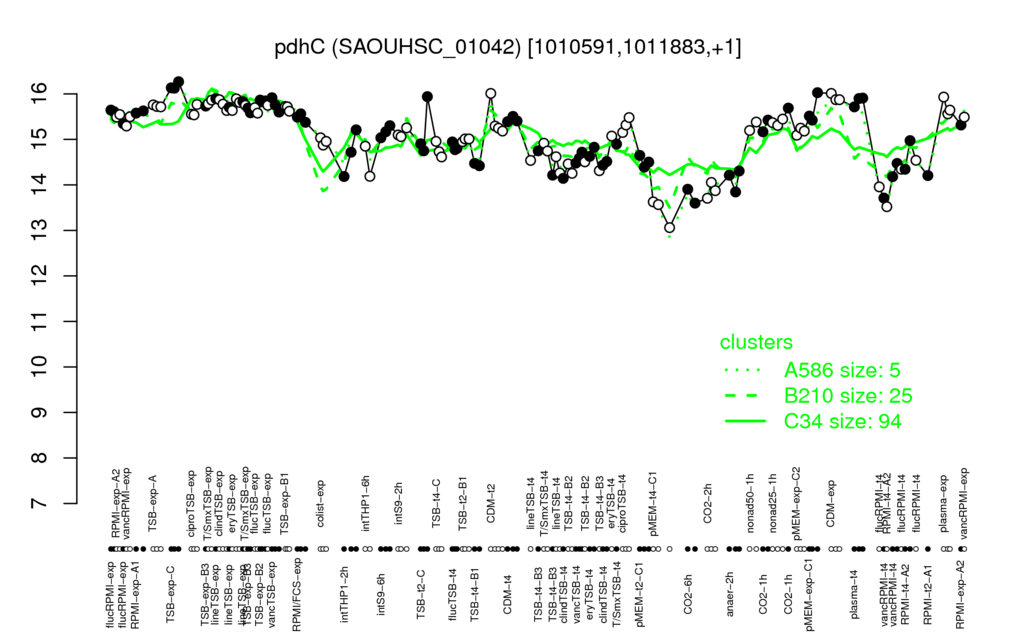
| TSB-exp-C | CDM-exp | TSB-exp-B2 | plasma-t4 | lineTSB-exp |
| 16.2 | 15.9 | 15.8 | 15.8 | 15.8 |
| pMEM-t4-C1 | CO2-6h | CO2-2h | anaer-2h | vancRPMI-t4 |
| 13.4 | 13.8 | 13.9 | 14.1 | 14.1 |
| pdhB | pdhD | pdhA | S426 | mraY | S610 | SAOUHSC_01283 | fabD | fabG | rsmI |
| 0.95 | 0.93 | 0.93 | 0.89 | 0.8 | 0.8 | 0.8 | 0.8 | 0.79 | 0.78 |
| SAOUHSC_02112 | S210 | S316 | SAOUHSC_02827 | S7 | S211 | S243 | SAOUHSC_02334 | SAOUHSC_00962 | S831 |
| -0.74 | -0.7 | -0.7 | -0.69 | -0.69 | -0.68 | -0.68 | -0.66 | -0.66 | -0.66 |
