| Supplementary data of the associated publication Start Page |
|
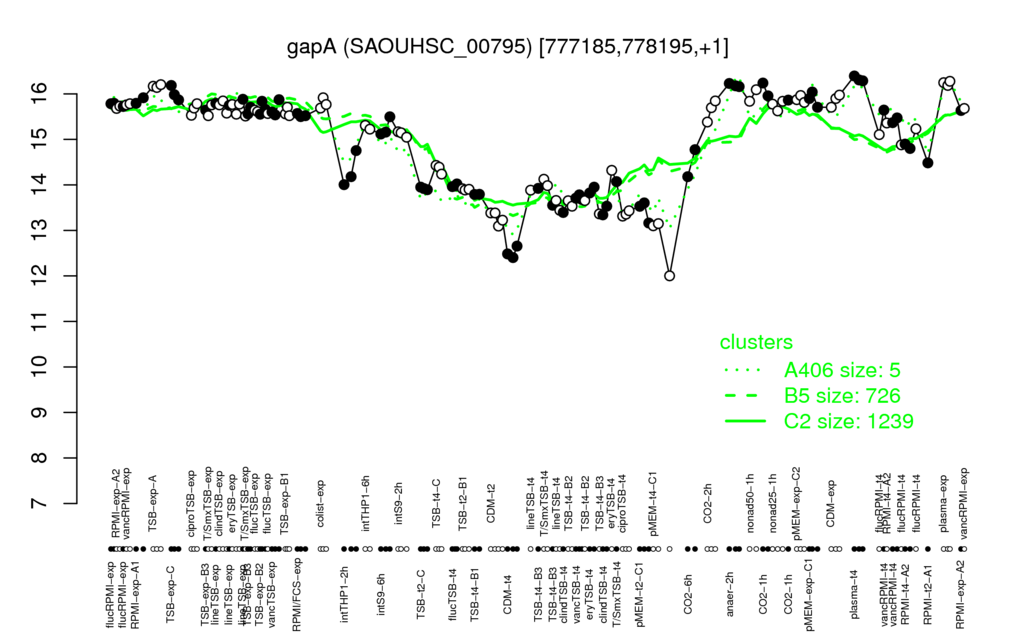
| plasma-t4 | plasma-exp | anaer-2h | TSB-exp-A | CO2-1h |
| 16.3 | 16.2 | 16.2 | 16.2 | 16 |
| CDM-t4 | pMEM-t4-C1 | CDM-t2 | ciproTSB-t4 | clindTSB-t4 |
| 12.5 | 12.8 | 13.3 | 13.4 | 13.4 |
| gapR | S322 | S323 | SAOUHSC_00462 | proS | S970 | pgk | SAOUHSC_01239 | S279 | metS |
| 0.96 | 0.96 | 0.91 | 0.9 | 0.89 | 0.88 | 0.88 | 0.88 | 0.88 | 0.87 |
| hutU | S1010 | SAOUHSC_02425 | hutI | SAOUHSC_02868 | rocF | gapB | putA | SAOUHSC_02376 | S593 |
| -0.93 | -0.93 | -0.92 | -0.92 | -0.92 | -0.92 | -0.91 | -0.91 | -0.9 | -0.9 |
