MuGeN is a software package for the visual exploration of multiple annotated genome portions. It is capable of simultaneously displaying genome portions loaded from various sources both local and remote and mix these with analysis result plots. It can also be used to generate images of these displays in a wide range of formats (PNG, PostScript, IMAP, XFig). Analysis results conform to an XML DTD, allowing easy integration of third-party analysis packages. Most of the display parameters can be customized, either interactively or through a preferences file.
(Click to enlarge.)
|
|
|
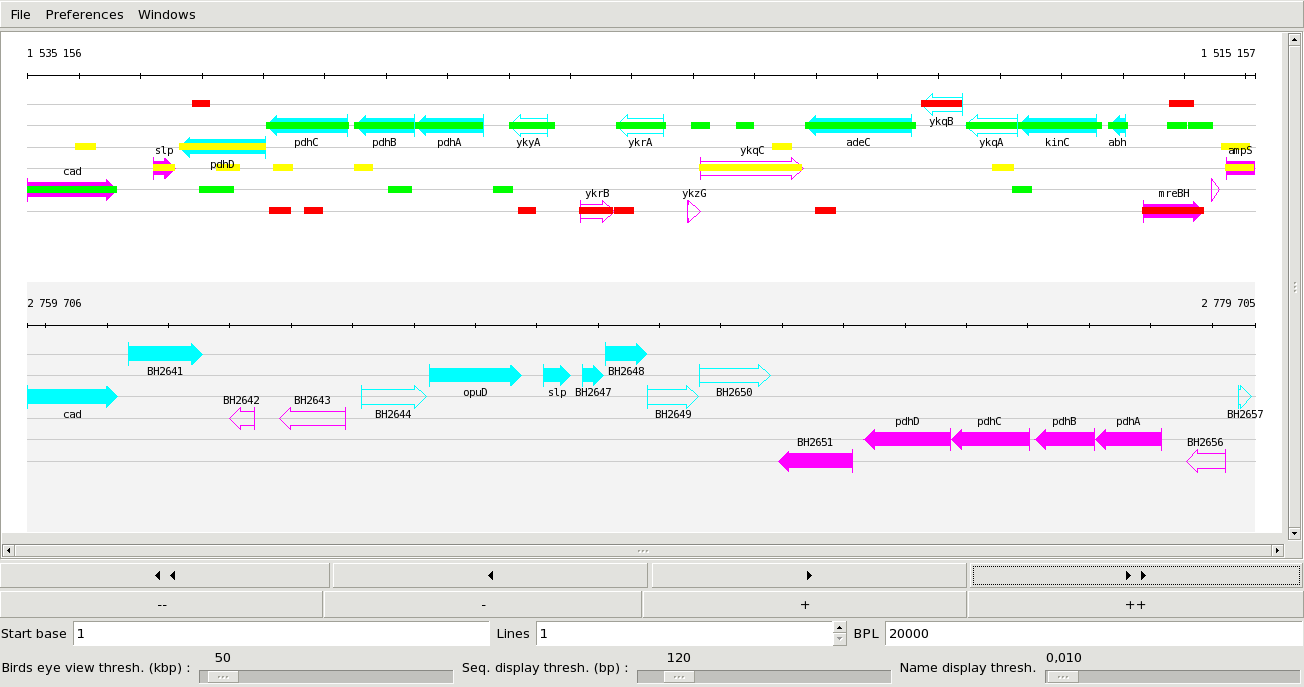
The main display window showing a portion of 20 kb of the genomes of Bacillus subtilis and B. halodurans. Widgets at the bottom of the window allow navigating through the genomes and various other display settings.
|
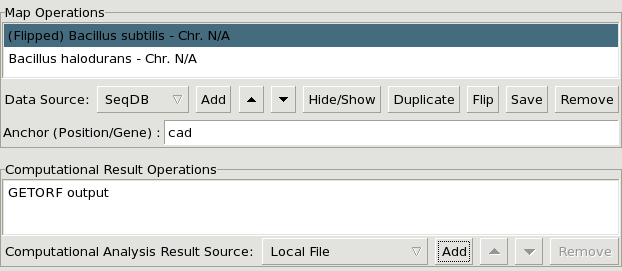
The list window displayig the currently loaded map entries and their associated analysis results.
|
|

|
|
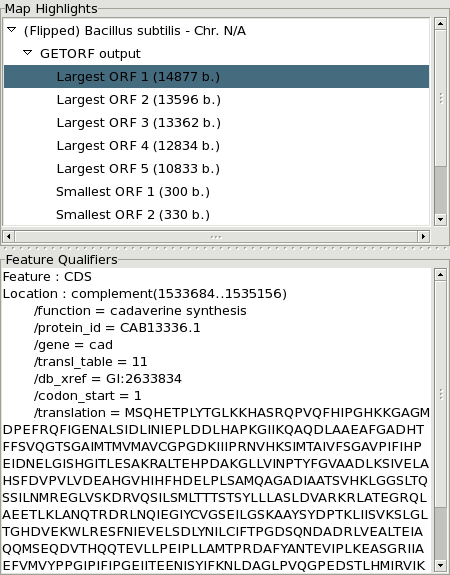
The information window giving the list of highlights associated with the analysis results as well as the list of qualifiers and values of the feature currently under the mouse pointer.
|
The result information window giving more details about computational analysis results. Its content may be saved to a text file.
|

|
First 100 kb portions of the genomes of Listeria monocytogenes, L. innocua and Bacillus halodurans, each displayed with a different background color.
|
|
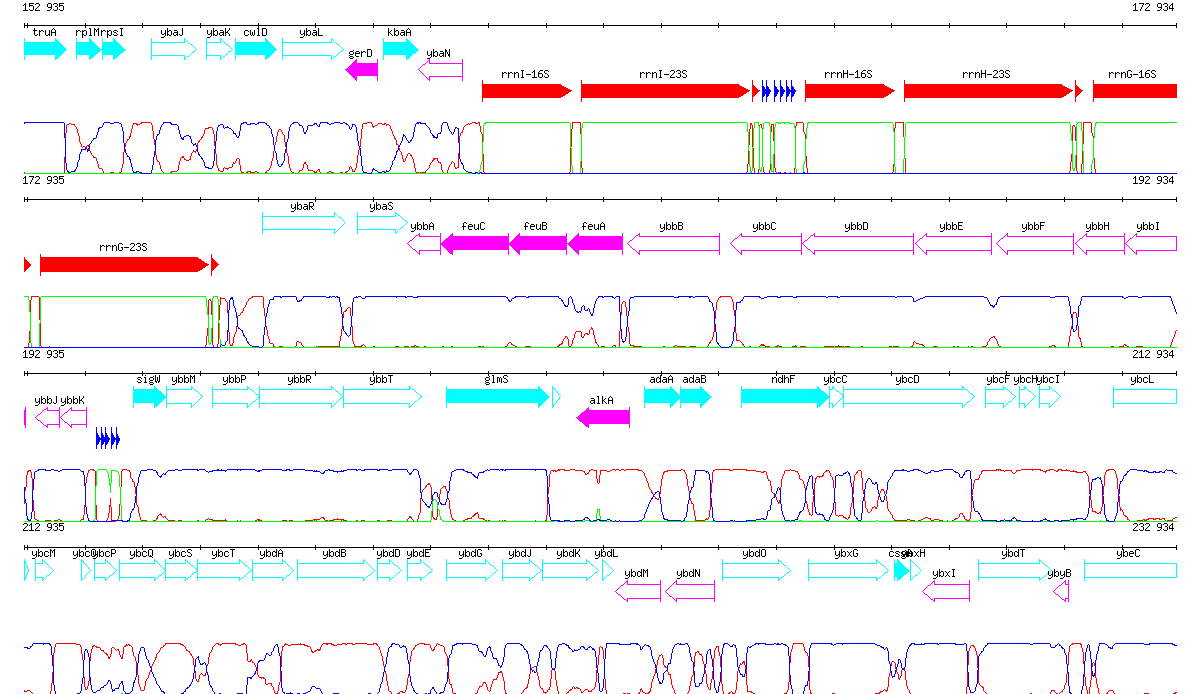
80 kb portion of the genome of Bacillus subtilis starting with gene truA. The lineplot shows the segmentation of the chromosome into 3 states according to the RHOM program.
|
|
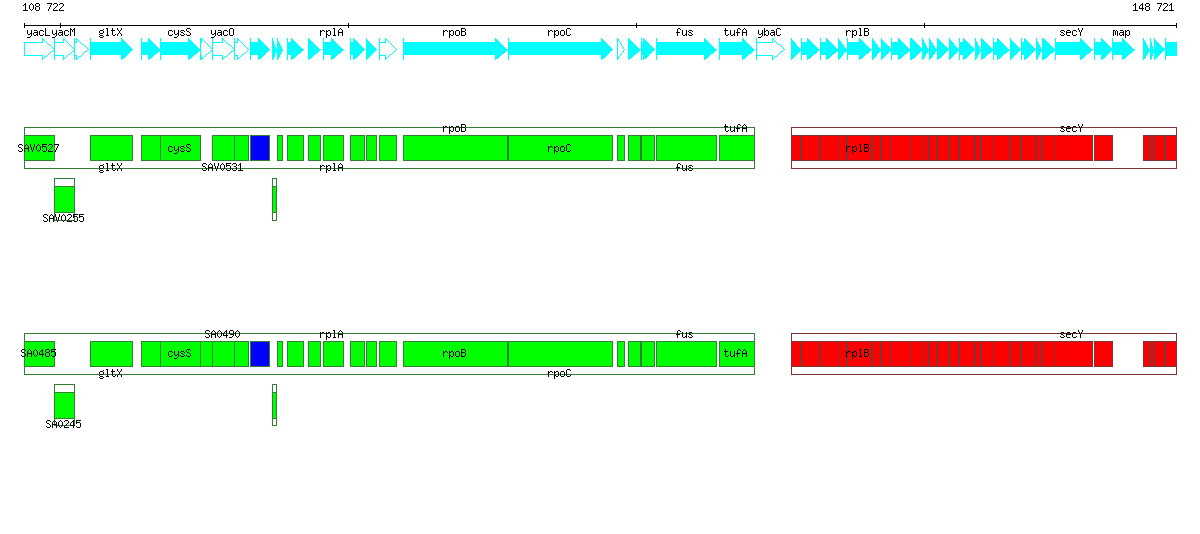
40 kb portion of the genome of B. subtilis showing its conservation in the genomes of L. innocua and L. monocytogenes. Box plots below the feature line show "alignemnts" of conserved orthologs and/or homologs. Green (resp. blue) boxes denote orthologs (resp. homologs) in a conserved portion on identical strands. Red boxes denote orthologs in a conserved portion on the opposite strand.
|
|
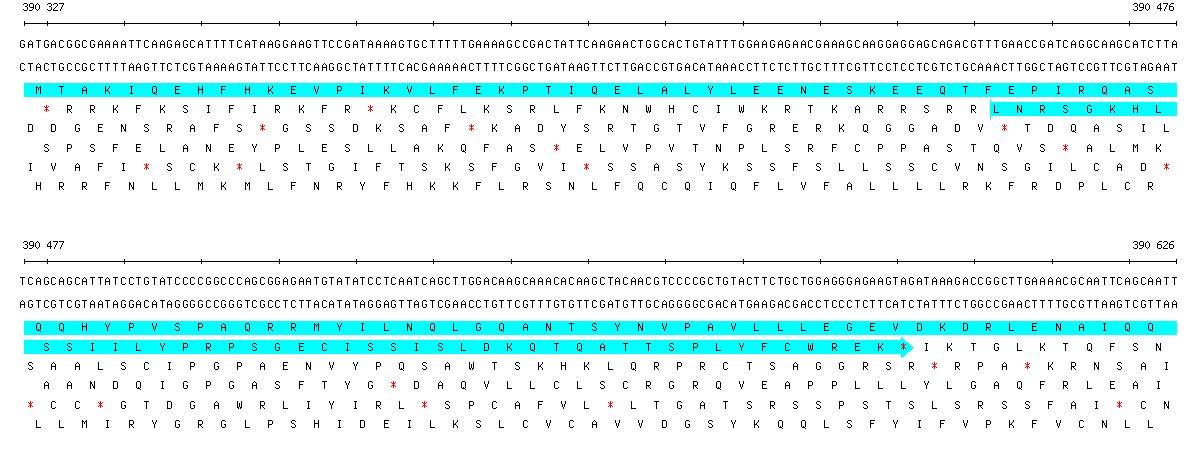
Sequence view of a 300 bp portion of B. subtilis showing two overlapping CDS annotations.
|
|
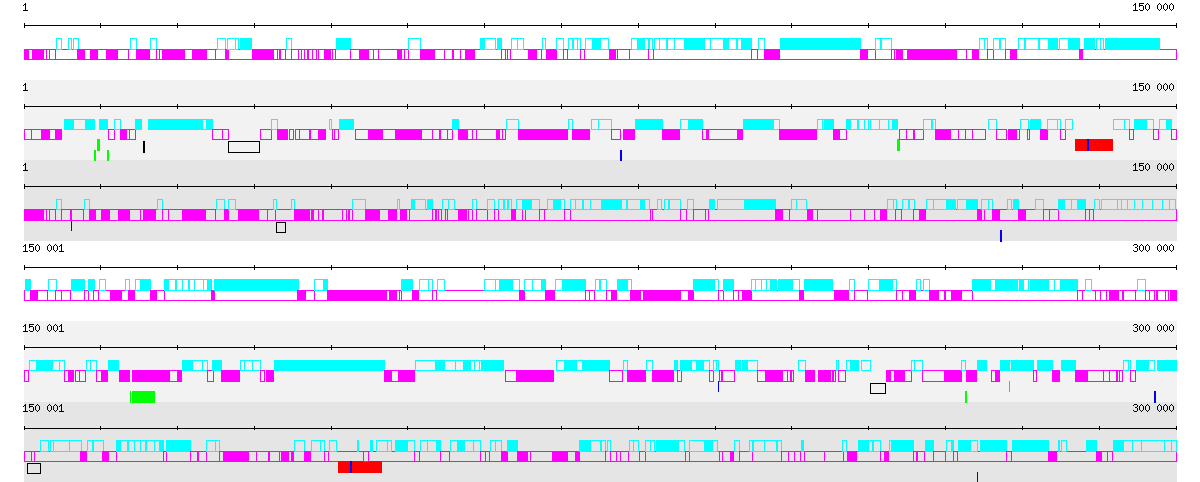
Birdseye view of the first 300 kb of the genomes of Pyrococcus abyssi, P. furiosus and P. horikoshii.
|
|
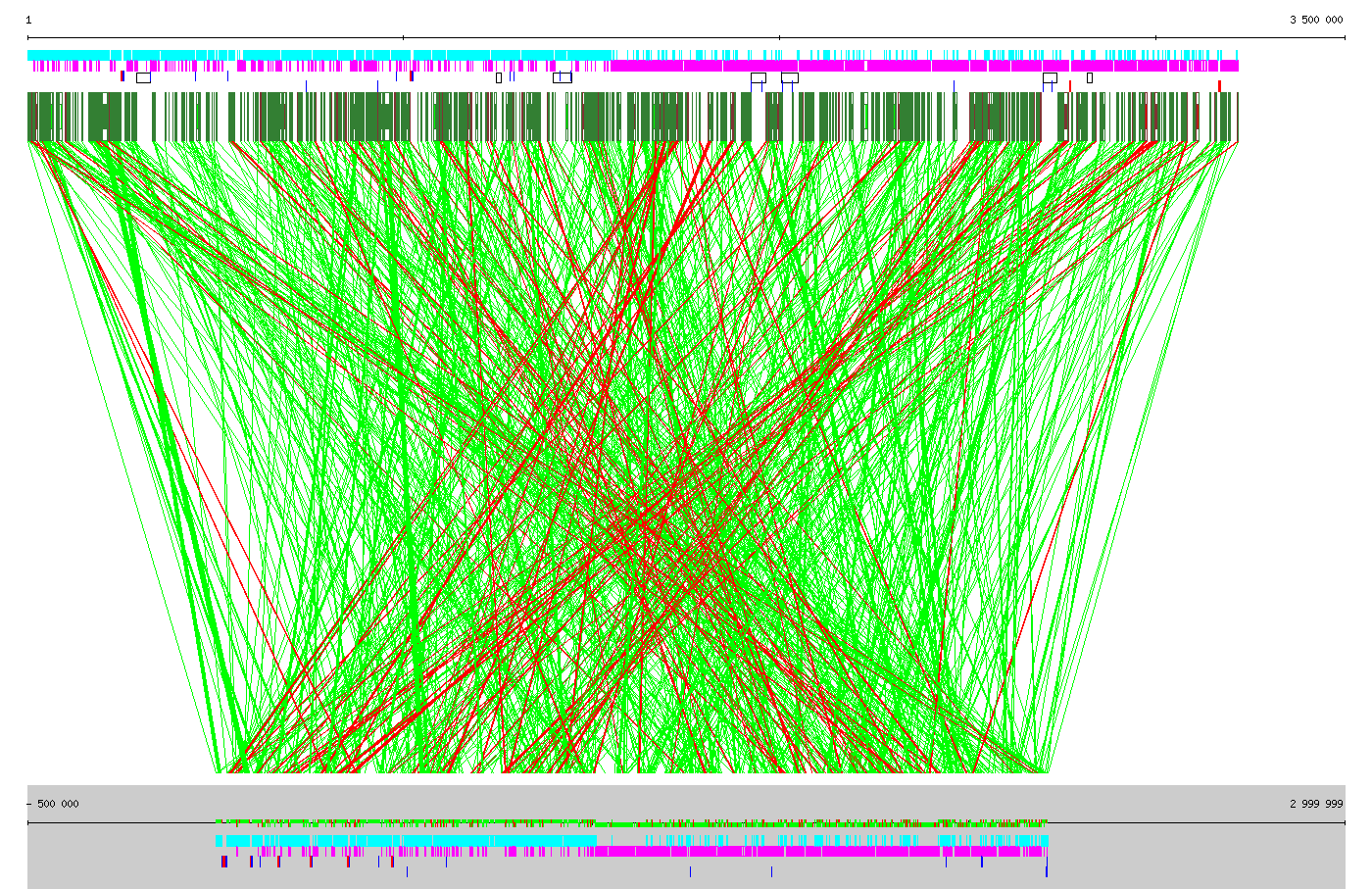
Birdseye view of the complete genome maps of Enterococcus faecalis V583 (top) and Streptococcus agalactiae NEM316 (bottom). Boxes below the annotations of E. faecalis denote genes present in both organisms. Links show the genes present in both organisms and located in conserved genomic contexts of at least 3 consecutive geens. Green links stand for conserved context of identical direction in the two organisms. Red links stand for inverted conserved contexts.
|

|
View of a conserved gene context present in both E. faecalis and S. agalactiae. Genes bkdA, bkdB and bkdC form a conserved gene context of opposite direction in both organisms (shown by the red links). Gene bkdD is also present in the same neighborhood though it's precise location differs in the two organisms.
|
MuGen is bundled in two separate archives: the first one contains the set of Perl scripts and modules needed to run MuGeN; the second one contains sample data illustrating various features of MuGen.
MuGeN is now hosted on MIA's forge at http://mulcyber.toulouse.inra.fr/projects/mugen. The latest distribution(s) are no longer listed below but can be downloaded from MIA's forge.
Older available releases are :
The documentation is alos hosted at MIA's forge here.
The following list gives a set of applications relying on MuGeN for data visualiszation
- Micado, the Microbial Advanced DataBase Organization uses MuGeN to generate maps of genome portions that are included in Web pages.
- SHOW generates probability profiles which can be superposed on Genome Maps
- Origami the Online Resource of Inegrated Genomes and Analysis results for MIcrobes offers a set of completely sequenced genomes together with analysis results about comparative genomics.
- MOSAIC, an INRA in house project carrying out comparative genomics on Escherichia coli strains.
MuGeN has been the subject of the paper with the following reference:
MuGeN: simultaneous exploration of multiple genomes and analysis results
M. Hoebeke, P. Nicolas and P. Bessières
Bioinformatics 19(7): 859-864
Mark Hoebeke
Unité Statistique & Génome - Tour Evry 2
523, place des Terrasses
F - 91000 Evry
FRANCE
Mark.Hoebeke_AT_jouy.inra.fr
|










